
|
 |
 |
Newsletter 01/2022
|
 |
 |
XCyting New Break Apart Probes…We are very pleased to announce three new XCyting locus-specific break apart probes in addition to our hematology and oncology portfolio. XL TCL1 BA is designed to detect TCL1 gene cluster locus involving rearrangements described in several T-cell prolymphocytic leukemia (T-PLL) cases, which are rare but aggressive T-cell malignancies. XL SPI1 BA is suitable for detection of the SPI1 gene locus involving rearrangements, which occur in 3,9% of all pediatric T-cell acute lymphocytic leukemia (T-ALL) cases. Known fusion partners of SPI1 are the genes TCF7, STMN1, BCL11B. XL CSF1R BA is an extension of our BCR-ABL1-like ALL portfolio. CSF1R is one of the prominent genes involved in multiple rearrangements, that have been described in this high-risk subset of ALL disorders, which account for approximately 25% of all B-cell precursor ALLs.
|
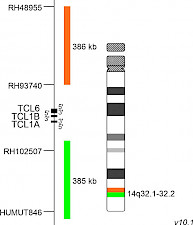
XL TCL1 BAD-5142-100-OG Clinical Applications: T-PLLThe gene TCL1A (TCL1 family AKT coactivator A, previously known as T-cell leukaemia/lymphoma 1A), has been described first in the early 1990s. It belongs to the TCL1 gene family including TCL1A (14q32.13), TCL1B (14q32.13) and MTCP1 (Xq28). Physiologically, the TCL1-encoded protein is expressed in fetal tissues and during early developmental stages of lymphocytes. It regulates many proteins responsible for cellular proliferation, survival and epigenetic modifications through multiple signalling pathways. Dysregulated TCL1 expression levels have been detected in chronic lymphocytic leukaemia (CLL), various lymphomas and in T-cell prolymphocytic leukaemia (T-PLL). In T-cells, TCL1 dysregulation is caused by chromosomal rearrangement events bringing TCL1 under the control of T-cell receptor (TCR) enhancer elements. Two distinct breakpoint clusters have been described, which are located on both sides of a 160kb region flanking the TCL1A/TCL1B/TCL6 gene cluster. The rearrangements leading to TCL1A translocations with TCRα/TCRδ and TCRβ gene loci have been characterized precisely. The underlying chromosomal rearrangements are inv(14)(q11q32), being the most common aberration, t(14;14)(q11;q32.1) and t(7;14)(q35;q32.1). Phage display technology- and structure-based drug design approaches are used to target TCL1, due to its major role in the activation of different survival and proliferation maintaining pathways. XL TCL1 BA consists of an orange-labeled probe hybridizing proximal to the TCL1A/TCL1B gene region at 14q32.1 and a green-labeled probe hybridizing distal to the TCL1A/TCL1B gene region at 14q32.1-14q32.2. Download fact sheet (PDF) for further information.
|
|
 |

|
 |

|
|
|
 |
|
 |
|
 |
 |
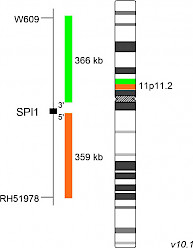
XL SPI1 BAD-5145-100-OG Clinical Applications: ALLThe SPI1 (SFFV provirus integration site-1) gene encodes the ETS-family transcription factor PU.1, which mediates gene expression during normal development of hematopoietic stem cells. SPI1 gene fusions were detected in 7 of 181 (3.9%) pediatric T-cell acute lymphoblastic leukemia (T-ALL) cases, analyzed and described by Seki et al. Genetic consequences of chromosomal rearrangements involving SPI1 are gene fusions containing 3’ exons of SPI1 and 5’ portions of TCF7, STMN1 and BCL11B. The c-terminal DNA binding domain (ETS domain) of the PU.1 protein is present in all known fusions, irrespective of the fusion partner. Fusion-positive samples show strongly increased SPI1 expression levels, as the rearranged portion of SPI1 is placed under the control of heterologous promoters of the above-mentioned genes. As SPI1 is normally expressed as an early phase gene in T-cell development, expression of wildtype SPI1 or SPI1 fusion genes at later stages results in higher cell proliferation and differentiation/maturation blockade during T-cell development. Fusion-positive cases show significantly shorter overall survival and are incurable when treated with standard chemotherapy. SPI1 has recently been found to interact with the PML‐RARα complex. Additionally, SPI1 itself is transcriptionally regulated by the PML‐RARα fusion protein. XL SPI1 BA consists of an orange-labeled probe hybridizing proximal to the SPI1 gene region at 11p11.2 and a green-labeled probe hybridizing distal to the SPI1 gene region at 11p11.2. Download fact sheet (PDF) for further information.
|
|
 |

|
 |

|
|
|
 |
|
 |
|
 |
 |
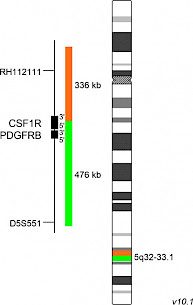
XL CSF1R BAD-5152-100-OG Clinical Applications: ALLIn the 2016 World Health Organization classification of myeloid neoplasms and acute leukemia, the B-lymphoblastic leukemia/lymphoma subtype ‘BCR-ABL1-like acute lymphoblastic leukemia’ (BCR-ABL1-like ALL), also called ‘Philadelphia chromosome (Ph)-like ALL’, was recognized as a provisional entity. BCR-ABL1-like ALL is a high-risk subset of B-cell precursor ALL disorders characterized by a highly similar gene expression profile compared to Ph+ ALL in the absence of BCR-ABL1 fusions. Altered genes involved in the development and maintenance of Ph-like ALL are ABL1, ABL2, BLNK, CRLF2, CSF1R, DGKH, EPOR, FGFR1, IL-2RB, JAK2, LYN, NTRK3, PDGFRA, PDGFRB, PTK2B and TYK2. CSF1R (colony stimulating factor 1 receptor) is a membrane-integrated tyrosine kinase receptor expressed by monocytes, macrophages, and other cells of the myeloid lineage. Binding of its ligands interleukin 34 or colony stimulating factor 1, respectively, leads to receptor homodimerization and auto-phosphorylation of the intracellular tyrosine kinase domains, subsequent signaling, and gene transcription. CSF1R-mediated signaling plays an important role in the differentiation and survival of monocytes and macrophages. In addition, CSF1R is a key factor in the tumor-permissive and immunosuppressive behavior of ‘tumor associated macrophages’ by promoting tumor progression and survival via suppressing effector T-cell functions. Known translocation partner genes involved in CSF1R rearrangements, identified in Ph-like cases, are SSBP2, MEF2D and TBL1XR1. The most intensively characterized CSF1R fusion is SSBP2-CSF1R resulting from the translocation t(5;5)(q14;q32). The ABL1 inhibitors Imatinib and Dasatinib are known to have anti-survival and pro-apoptotic capacities on cells expressing the SSBP2-CSF1R fusion. XL CSF1R BA consists of an orange-labeled probe hybridizing proximal to the CSF1R gene region at 5q32 extending into the gene up to intron 5 and a green-labeled probe hybridizing distal to the CSF1R gene region at 5q32-33.1 extending into the gene up to intron 5 (GRCh37/hg19). Download fact sheet (PDF) for further information.
|
|
 |

|
 |

|
|
|
 |
|
 |
|
|
 |
 |

|
|