|
View in web browser
|

|
 |
 |
Newsletter 01/2023
|
 |
 |
XCyting New MetaSystems Probes…We are very pleased to announce four new XCyting locus-specific probes expanding our hematology and microdeletion portfolio. XL t(11;14) CCND1/IGH DF is designed as an alternative variant of XL t(11;14) MYEOV/IGH DF (D-5111-100-OG), in which the MYEOV locus is not flanked by the orange-labeled probe. Translocations of the type t(11;14)(q13;q32) are described in non-hodgkin lymphoma (NHL), chronic lymphocytic leukemia (CLL), and multiple myeloma (MM) cases. XL CCND3/IGH DF is a further developed version of XL t(6;14) CCND3/IGH DF (D-5109-100-OG) introducing a new, the entire CCND3 gene covering design. t(6;14)(p21;q32) resulting in CCND3 overexpression by juxtaposing the CCND3 gene with enhancer elements of the IGH locus. Translocation of the type t(6;14)(p21;q32) are recurrent translocation found in 4-5% of MM cases. XL CUX1/EZH2/7cen is designed as an additional option to the XL 7q22/7q36 (D-5043-100-TC) and XL del(7)(q22q31) D-5068-100-TC for the analysis of deletions in 7q22. The probe is intended to detect partial deletions of the long arm of chromosome 7 or monosomy 7. Both are highly recurrent chromosomal aberrations frequently observed in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). XL DiGeorge TBX1 is newly designed and the latest family member of our microdeletion probe portfolio. FISH probes detecting Wolf-Hirschhorn, Cri-du-Chat, Williams-Beuren, Prader-Willi/Angelman, and Smith-Magenis/Miller-Dieker syndromes have already been available since 2019 and 2020, respectively.
|
 |
 |
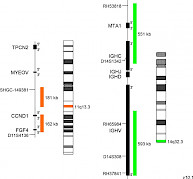
XL t(11;14) CCND1/IGH DFD-5140-100-OG Clinical Applications: NHL, CLL, MMMantle cell lymphoma (MCL) is a rare subtype of B-cell non-Hodgkin lymphoma (NHL) with an aggressive clinical course. MCLs represent 6-8% of all NHLs. However, 95% of all MCLs are positive for the translocation t(11;14)(q13;q32). To a lower extent, t(11;14)(q13;q32) has also been detected in approx. 20% of B-cell prolymphocytic leukemias, 3-5% of plasma cell myelomas/ multiple myelomas (MM), and in rare cases (<1%) of B-cell chronic lymphocytic leukemia (CLL). The breakpoints within the IGH locus are different in MM and MCL genomes containing translocation t(11;14). The translocation results in juxtaposition of the immunoglobulin heavy-chain (IgH) locus with the cyclin D1 (CCND1) gene locus, which is not transcribed in normal B-cells. As a result, cis IgH enhancer elements take over the transcriptional control of CCND1 leading to deregulated CCND1 expression (e.g. overexpression in MCL). CCND1 coordinates growth signals with cell cycle progression playing a key role in the regulation of the G1 to S phase transition. Additionally, it was shown that some oncogenic effects of t(11;14)(q13;q32) are not directly attributable to the deregulated CCND1 expression, but rather to epigenetic downstream mechanisms such as CCND1 locus methylation and histone acetylation. Since MCL is an aggressive B-cell neoplasm with a median survival of 3–5 years and has a poor response to traditional chemotherapeutic approaches, distinction of MCL from other lymphoproliferative disorders by differential diagnosis is of great importance. In multiple myeloma (MM), t(11;14) is the most common translocation, detectable by FISH in about 15-20% of all MM patients. Conventional cytogenetics has a much lower sensitivity, detecting t(11;14) in about 5% of MM patients. XL t(11;14) CCND1/IGH DF is designed as an alternative probe variant of XL t(11;14) MYEOV/IGH DF (D-5111-100-OG) resulting from the request of some professional users for a MYEOV gene locus free CCND1 probe part. XL t(11;14) CCND1/IGH DF consists of an orange-labeled probe hybridizing to the CCND1 gene region at 11q13.3 and a green-labeled probe hybridizing to the IGH gene region at 14q32.3.
|
|
 |

|
 |

|
|
|
 |
|
 |
|
 |
 |
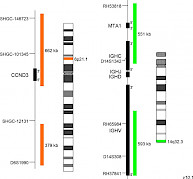
XL CCND3/IGH DF D-5147-100-OG Clinical Applications: MMThe most frequent primary abnormalities in multiple myeloma (MM) are trisomies of odd-numbered chromosomes or translocations involving the immunoglobulin heavy chain (IGH) gene locus. The most common MM-associated IGH translocations are t(11;14), t(4;14), t(6;14), t(14;16) and t(14;20) in the order of their occurrence. The consequence of these rearrangements is the dysregulation of genes juxtaposed to transcriptional enhancer elements in the IGH gene locus. Prognosis and risk stratification strongly depend on the detection and interpretation of cytogenetic primary abnormalities. Translocations t(14;16) and t(14;20) are considered as high risk, t(4;14) as intermediate risk and t(6;14) and t(11;14) as standard risk cytogenetic aberrations in patients with MM based on FISH testing. Secondary aberrations are also influencing the outcome. Cyclins of the D-family are essential for the transition from G1 to S phase during the cell cycle progression. t(6;14)(p21;q32) moves the cyclin D3 (CCND3) gene in proximity to 3’ IGH enhancer sequences and is associated with CCND3 overexpression. The chromosomal translocation has been reported as a rare but recurrent event not only in myeloma but also in other B-cell malignancies such as diffuse large B-cell lymphoma. XL CCND3/IGH DF is designed as an further developed variant of XL t(6;14) CCND3/IGH DF (D-5109-100-OG) featuring a new, the entire CCND3 gene covering design. XL CCND3/IGH DF consists of an orange-labeled probe hybridizing to the CCND3 gene and flanking regions at 6p21.1 and a green-labeled probe hybridizing to the IGH gene region at 14q32.3.
|
|
 |

|
 |

|
|
|
 |
|
 |
|
 |
 |
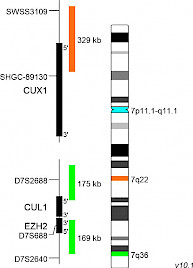
XL CUX1/EZH2/7cenD-5144-100-TC Clinical Applications: MDS, AMLPartial deletions of the long arm of chromosome 7 [del(7q),7q-] or monosomy 7 (-7) are highly recurrent chromosomal aberrations frequently observed in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). In MDS and AML, del(7q) as sole cytogenetic aberration has a prevalence of less than 5%. However, many of the examined patients with del(7q) have additional cytogenetic aberrations. Furthermore, whole-exome sequencing studies analyzed the coincidence of mutated genes (e.g. CUX1, LUCL2, CUL1, and EZH2) and -7 or del(7q). 7q deletions are assigned to the intermediate genetic prognostic risk group (ELN risk stratification) or the high risk prognostic group (revised MRC classification), in AML. However, 7q- are classified by the revised IPSS (International prognostic scoring system) as “intermediate cytogenetic prognostic risk” or “poor risk” - if coincide with another abnormality, in MDS. Several deleted regions have been identified along the long arm of chromosome 7, but two critical genomic regions are known to be commonly deleted regions (CDRs): 7q22 and 7q31-q36. Studies focusing on CDRs on 7q have highlighted the significance of the gene loci of EZH2, SAMD9L, CUX1, MLL3, and DOCK4, whose assumed roles as tumor suppressor genes could explain the criticality of their haploinsufficiency. The CUX1 gene locus has been described as frequently deleted locus in case of 7q deletions, increasing the accuracy of the diagnosis of 7q22 deletions. XL CUX1/EZH2/7cen (D-5144-100-TC) is designed as an additional option to the XL 7q22/7q36 (D-5043-100-TC) and XL del(7)(q22q31) D-5068-100-TC for the analysis of deletions in 7q22. The KMT2E gene region hybridizing probe portion at 7q22 is replaced by a partial CUX1 gene locus covering probe part. XL CUX1/EZH2/7cen consists of an orange-labeled probe hybridizing to the CUX1 gene region at 7q22, a green-labeled probe hybridizing to the EZH2 gene region at 7q36 and an aqua-labeled probe hybridizing to the centromere of chromosome 7.
|
|
 |

|
 |

|
|
|
 |
|
 |
|
 |
 |
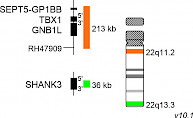
XL DiGeorge TBX1D-5415-050-OG Clinical Applications: Microdeletion Syndrome (MicroDel)The 22q11.2 deletion syndrome (22q11.2DS) includes the DiGeorge- and Velo-cardio-facial syndrome and is the most common microdeletion syndrome. Up to 95% of the deletions identified occur de novo and are not inherited. 22q11.2DS related de novo deletions are mediated by non-allelic inter- and intrachromosomal homologous recombination between low copy number repeats on chromosome 22. A deletion of 3 Mb overlapping about 90 genes, is detected in the vast majority of 22q11.2DS cases whereas a smaller fraction is identified with a nested deletion of about 1.5 Mb overlapping with about 55 genes. One of the most thoroughly investigated genes in this region is the T-box transcription factor 1 (TBX1). TBX1 is phylogenetically conserved and has manifold functions during embryonic development. Studies using mouse as a model organism have shown that TBX1 is a key determinant in 22q11.2DS. XL DiGeorge TBX1 consists of an orange-labeled probe hybridizing to the TBX1 gene region at 22q11.2 and a green-labeled probe hybridizing to the SHANK3 gene region at 22q13.3.
|
|
 |

|
 |

|
|
|
 |
|
 |
|
|
 |
 |

|
|
 |
|
|
 |
|