Probes Newsletter 05/2019
Newsletter 05/2019

Xmas is going to be XCyting…
We are proud to present four new XCyting locus-specific probes products intended for use in hematology and pathology. XL t(8;9) PCM1/JAK2 DF is a new probe completing our portfolio detecting chromosomal aberrations in the context of CML/MPN diseases. WHO defined this new entity in 2017 as “myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB or FGFR1, or with PCM1/JAK2”. XL IGH/MAFB DF is used for the detection of the multiple myeloma associated translocation t(14;20). It is designed as an improved variant of XL t(14;20) IGH/MAFB featuring a new, MAFB gene covering design. XL t(14;18) IGH/MALT1 DF is designed to detect MALT lymphoma associated t(14;18)(q32;q21). This probe is intended for methanol/acetic-acid fixed cells and tissue sections. XL ABL1 BA is a further member of our growing Philadelphia-like ALL panel and is a logical complement for established probes in the field of Philadelphia-like ALL, CML/MPN and AML. For further information, please visit www.metasystems-probes.com.
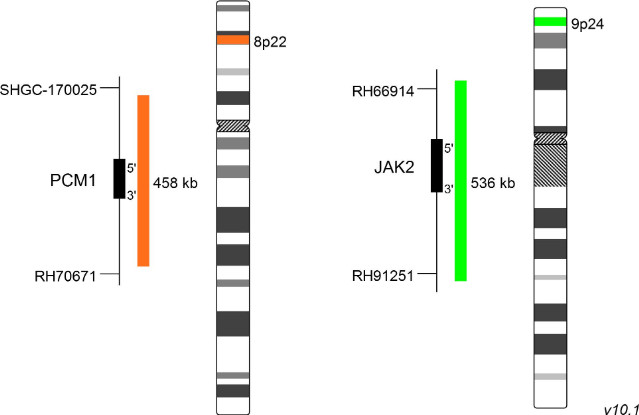
XL t(8;9) PCM1/JAK2 DF
Clinical Applications: CML/MPN
Receptor tyrosine kinases as well as Janus kinases (JAKs) have multiple functions in the signal transmission of cytokines and growth factors. Furthermore, constitutively enhanced JAK phosphorylation levels play critical roles in chronic inflammatory processes and cancer development. Chromosomal rearrangements involving JAK2, leading to constitutive activation of the kinase, have been detected in several patients with myeloproliferative neoplasm (MPN) or myelodysplastic syndrome (MDS). PCM1-JAK2 represents the most frequent JAK2 fusion gene. The responsible translocation generating the described fusion is t(8;9)(p22;p24.1). In 2017 the WHO updated the category "myeloid/lymphoid neoplasms with eosinophilia and gene rearrangement" by adding a provisional entity, PCM1-JAK2. As the presence of this rearrangement is associated with a poor prognosis, only allogeneic hematopoietic stem cell transplantation is considered to be rather successful, although first small studies using the JAK2 inhibitor ruxolitinib as treatment for patients with PCM1-JAK2 positive chronic eosinophilic leukemia, not otherwise specified, have shown promising short-term remission.
XL t(8;9) PCM1/JAK2 DF is designed as a dual fusion probe. The orange labeled probe spans the breakpoint at 8p22 (PCM1), and the green labeled probe spans the breakpoint at 9p24 (JAK2). Download fact sheet (PDF) for further information.
XL IGH/MAFB DF
Clinical Applications: MM
The most frequent primary abnormalities in multiple myeloma (MM) are trisomies of odd-numbered chromosomes or translocations involving the immunoglobulin heavy chain (IGH) gene locus. The most common MM-associated IGH translocations are t(11;14), t(4;14), t(6;14), t(14;16) and t(14;20) in the order of their occurrence. As a consequence, translocation partner genes of IGH are dysregulated, as they are juxtaposed to transcriptional enhancers in the IGH locus. Even if associated with poor prognosis in MM, MGUS/SMM cases characterized by the presence of t(14;20) can be stable for years before progression occurs, whereas MGUS/SMM cases with t(4;14) and t(14;16) show a significantly faster progression rate. The recurrent translocation t(14;20) (q32;q12) results in ectopic expression of the basic leucine zipper transcription factor MAFB which plays an important role in lineage-specific hematopoiesis. Furthermore, t(14;20) is associated with poor prognosis by promoting high cyclin D2 activity, thereby dysregulating normally balanced cell cycle.
XL IGH/MAFB DF is designed as an improved variant of XL t(14;20) IGH/MAFB DF (D-5105-100-OG) featuring a new, MAFB gene covering design.
XL IGH/MAFB DF probe is designed as a dual fusion probe. The orange labeled probe covers the MAFB gene and flanks the breakpoint at 20q12, the green labeled probe flanks the IGH breakpoint region at 14q32. Download fact sheet (PDF) for further information.
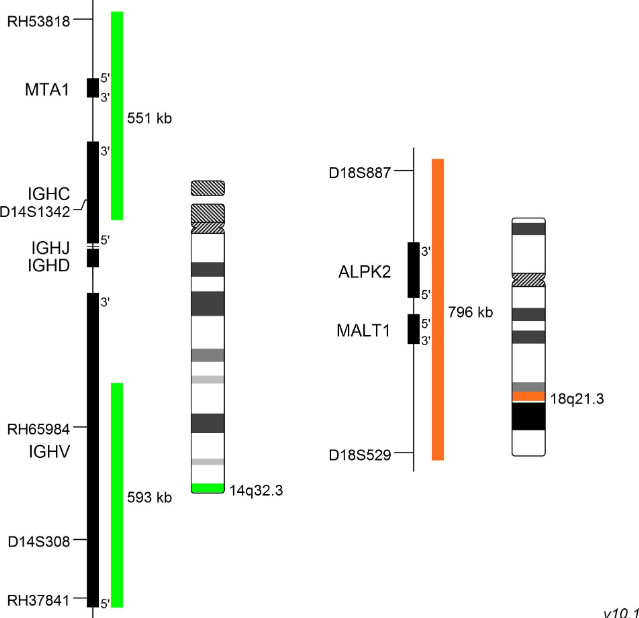
XL t(14;18) IGH/MALT1 DF
Clinical Applications: NHL
MALT (mucosa-associated lymphoid tissue) lymphomas occur at diverse anatomic sites and are closely linked to several distinct chronic inflammatory disorders. Up to 50% of the MALT lymphoma cases analyzed demonstrate MALT1 rearrangements. The MALT1 gene was originally identified by its involvement in the MALT lymphoma associated translocation t(11;18)(q21;q21). Approximately 20% of all MALT lymphoma cases are characterized by the presence of t(14;18)(q32;q21) leading to a IGH-MALT1 fusion. This reciprocal translocation juxtaposes MALT1 to transcriptional enhancers in the IGH locus and results in overexpression of the MALT1 gene. The distinct breakpoints on both chromosomes are precisely defined. The oncogenic potential of MALT1 is linked to its participation in the activation of nuclear factor-kappa B (NF-κB). This important transcription factor mediates the expression of anti-apoptotic, cell survival- and proliferation-promoting genes.
XL t(14;18) IGH/MALT1 DF is designed as a dual fusion probe. The orange labeled probe covers the MALT1 gene at 18q21, the green labeled probe flanks the IGH breakpoint region at 14q32. This probe is intended for methanol/acetic-acid fixed cells and tissue sections. Download fact sheet (PDF) for further information.
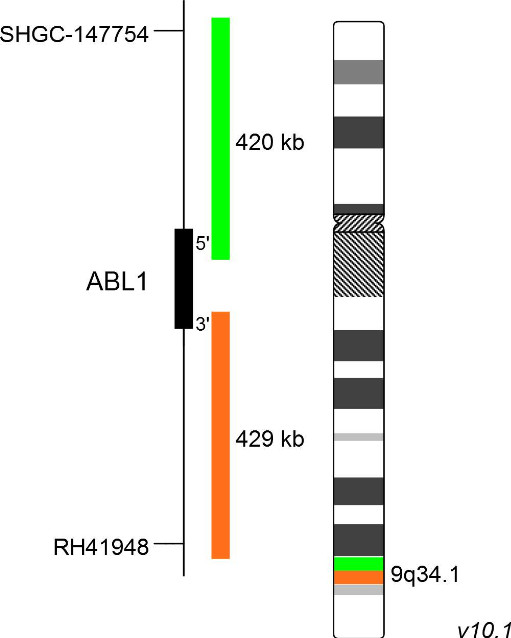
XL ABL1 BA
Clinical Applications: ALL, AML, CML/MPN
In 2017, the WHO recognized BCR-ABL1-like ALL, a subtype of B-lymphoblastic leukemia/lymphoma, as new entity. BCR-ABL1-like ALL, also known as Philadelphia chromosome (Ph)-like ALL, is found in 10-20% of childhood and in 20-30% of all adult B-cell dependent ALL cases. BCR-ABL1-like ALL is characterized by a gene expression profile sharing significant overlap with that of Ph-positive (Ph+) ALL. In contrast to Ph+ ALL, defined by the presence of the BCR-ABL1 fusion resulting from t(9;22)(q34;q11), BCR-ABL1-like ALL cases include a variety of genomic alterations enhancing kinase and cytokine receptor signaling. Prominent genes involved in the pathogenesis of BCR-ABL1-like ALL are CRLF2, EPOR, JAK2, ABL1, ABL2, CSF1R and PDGFRB. The already known heterogenous 5' fusion partners of ABL1 are CENPC, ETV6, FOXP1, LSM14A, NUP153, NUP214, RANBP2, RCSD1, SFPQ, SNX1, SNX2, SPTAN1 and ZMIZ1. The kinase domain of ABL1 is present in all identified chimeric fusion proteins.
XL ABL1 BA is designed as a break apart probe. The orange labeled probe hybridizes distal to the ABL1 gene region at 9q34.1 and extends into the gene up to intron 3, the green labeled probe hybridizes proximal to ABL1 and extends into the gene up to intron 1. (GRCh37/hg19). Download fact sheet (PDF) for further information.