Probes Newsletter 02/2019
Newsletter 02/2019

XCyting News!
We are very pleased to announce two new innovations in our XCyting Locus-Specific Probes product line for Hematology and Oncology. XL PDGFRA BA is a logical complement to our products XL 4q12 and XL 4q12 DC which are designed to detect the interstitial deletion of CHIC2 as a surrogate for the FIPL1-PDGFRA fusion gene. XL PDGFRA BA has a wider range of applications, detecting breaks in the PDGFRA gene locus. XL KMT2A BA has a design resembling that of the well proven probe XL MLL plus. However, the revised design of XL KMT2A BA allows the detection of cryptic insertions as an additional benefit.
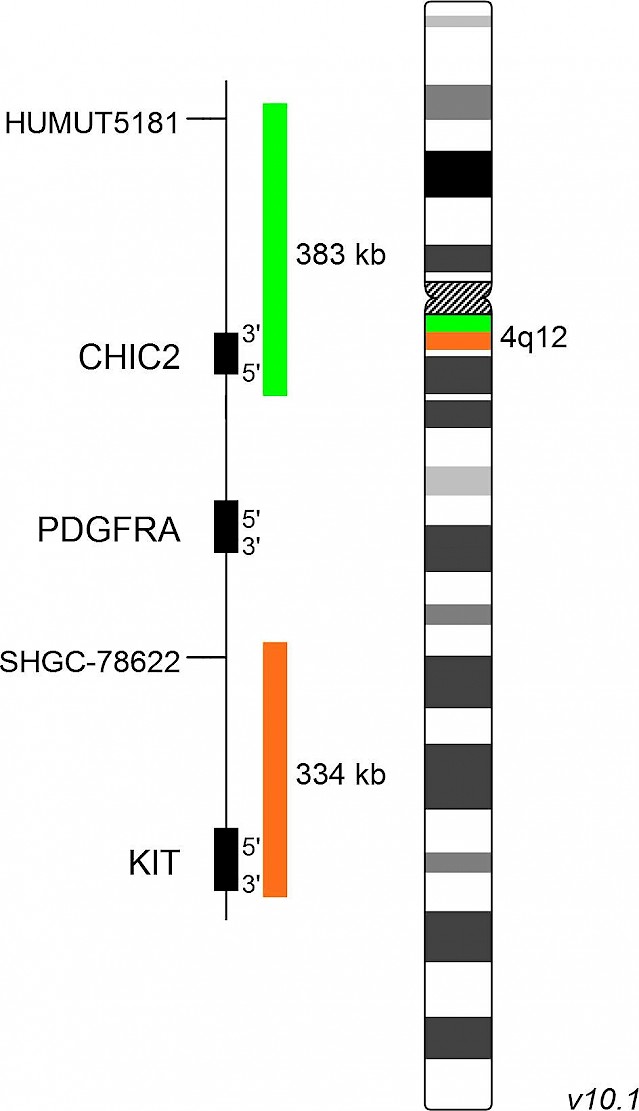
XL PDGFRA BA
Clinical Applications: CEL, AML, ALL
XL PDGFRA BA is designed as a break apart probe. The orange labeled probe hybridizes distal to the breakpoint in the PDGFRA gene region at 4q12, the green labeled probe hybridizes proximal to the breakpoint and covers the CHIC2 gene.
The category "myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB or FGFR1, or with PCM1/JAK2" of the revised 4th edition of the WHO classification defines three particular groups with rearrangements in PDGFRA, PDGFRB or FGFR1 and a provisional entity with PCM1/JAK2 rearrangement. Neoplasms with eosinophilia are associated with dysregulated tyrosine kinases, usually as a result of gene fusions. The most frequent PDGFRA-related aberration is the interstitial deletion of the CHIC2 gene with breakpoints in the FIP1L1 and PDGFRA genes. However, translocations with other partner genes, resulting in aberrant tyrosine kinase activity, are known. Involvement of PDGFRA can be detected by break apart strategies with FISH.
Download fact sheet (PDF) for further information
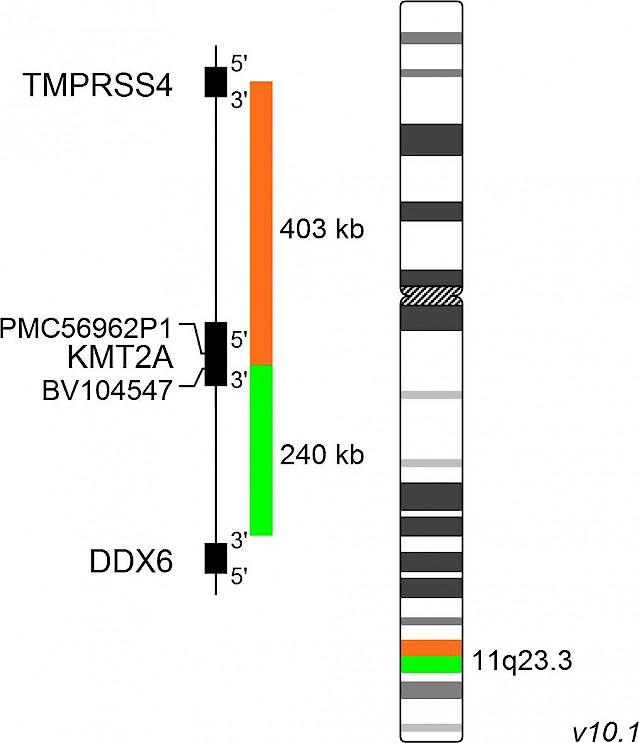
XL KMT2A BA
Clinical Applications: ALL, AML
XL KMT2A BA is designed as a break apart probe. The orange labeled probe hybridizes proximal to KMT2A at 11q23 and extends into the gene up to intron 24, the green labeled probe hybridizes distal to KMT2A and extends into the gene up to intron 20 and thus overlapping each other for 3.4kb (GRCh37/hg19).
The KMT2A (formerly MLL) gene, located on chromosome 11q23, is rearranged in about 10% of all acute leukemia patients. Today, more than 80 translocation partners of KMT2A have been identified. Translocations are resulting in in-frame fusions between the KMT2A part N-terminal to the break point cluster region and the respective fusion partners. Breaks within the KMT2A breakpoint cluster region result in a small orange split signal remaining with the separated green signal. The small orange signal might be visible in some cases. The design of XL KMT2A BA allows the detection of cryptic insertion of portions of KMT2A into other chromosomes indicated by the signal constellation 1O2GO.
Download fact sheet (PDF) for further information
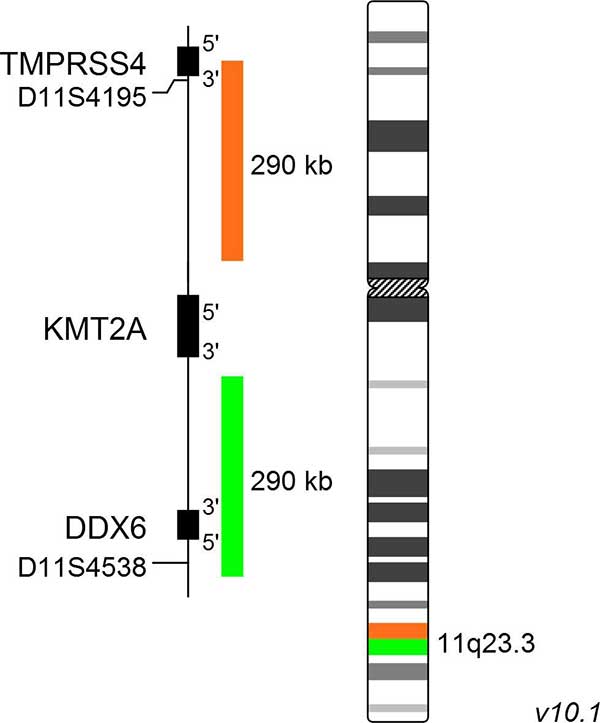
XL MLL plus
Clinical Applications: ALL, AML
The XL MLL plus probe is designed as a break apart probe. Its orange labeled part hybridizes proximal to the KMT2A (formerly MLL1) gene locus at 11q23, the green labeled probe hybridizes to the distal region. The probe does not cover coding regions within the KMT2A gene.
XL MLL plus will remain part of our portfolio for all customers who don´t want to switch to the new development XL KMT2A BA.