
XL t(12;21) ETV6/RUNX1 DF
Translocation/Dual Fusion Probe
- Order Number
- D-5115-100-OG
- Package Size
- 100 µl
- Regulatory Status
- IVDD
IVDR Certification
MetaSystems Probes has already certified a wide range of FISH probes, according to IVDR.
This product remains IVDD-certified until further notice.
Discover all IVDR-certified products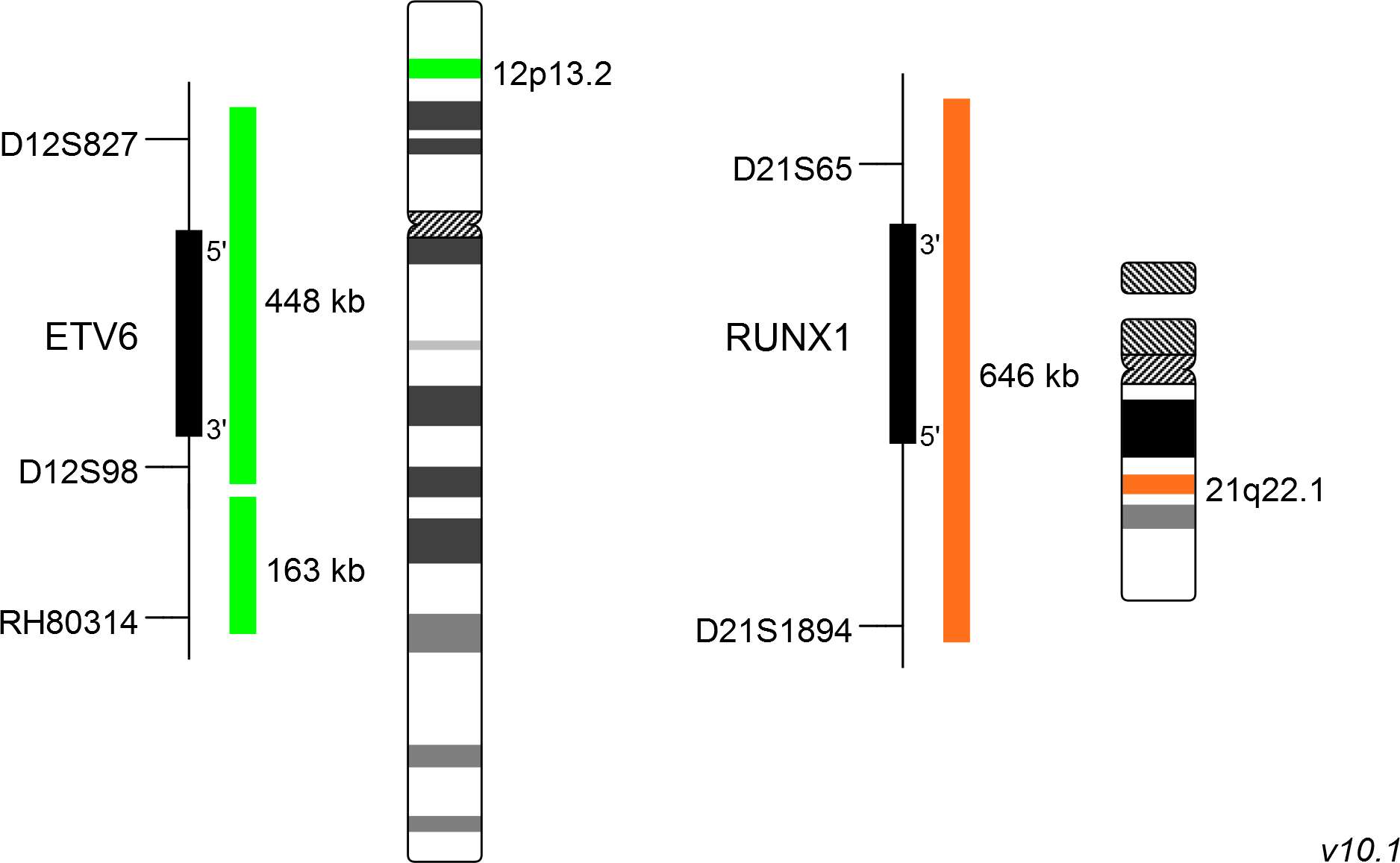
XL t(12;21) ETV6/RUNX1 DF consists of a green-labeled probe hybridizing to the ETV6 gene region at 12p13.2 and an orange-labeled probe hybridizing to the RUNX1 gene region at 21q22.1.
Probe maps for selected products have been updated. These updates ensure a consistent presentation of all gaps larger than 10 kb including adjustments to markers, genes, and related elements. This update does not affect the device characteristics or product composition. Please refer to the list to find out which products now include updated probe maps.
Probe map details are based on UCSC Genome Browser GRCh37/hg19, with map components not to scale.
Acute lymphoblastic leukemia (ALL) is a rapidly progressing cancer type characterized by the malignant transformation of lymphoid progenitor cells. It is the most common childhood cancer type and the second most common leukemia in adults. Treatment of children usually results in good prognosis whereas the outcome for adults is less optimistic. Most patients show a transformation of precursors of the B-cell type, but also the T-cell phenotype is frequently observed. The most common aberration in pediatric B-cell ALL is t(12;21)(p13;q22) with an incidence of about 25%, compared to <5% in adults. The result of this reciprocal translocation is an ETV6/RUNX1 fusion gene. Scientific data suggest that ETV6/RUNX1 is already established prenatally, but additional chromosomal aberrations are necessary for the development of ALL postnatally. The ETV6/RUNX1 fusion gene is transcriptionally active and is dysregulating a cascade of downstream genes. One study has shown, that all positive t(12;21) cases harbored the ETV6/RUNX1 fusion gene but not the reciprocal gene. This suggests, that ETV6/RUNX1 is involved in the manifestation of ALL, but not RUNX1/ETV6.
Since t(12;21) is not detectable by conventional cytogenetic methods, FISH is one of the methods of choice.
Clinical Applications
- Acute Lymphoblastic Leukemia (ALL)
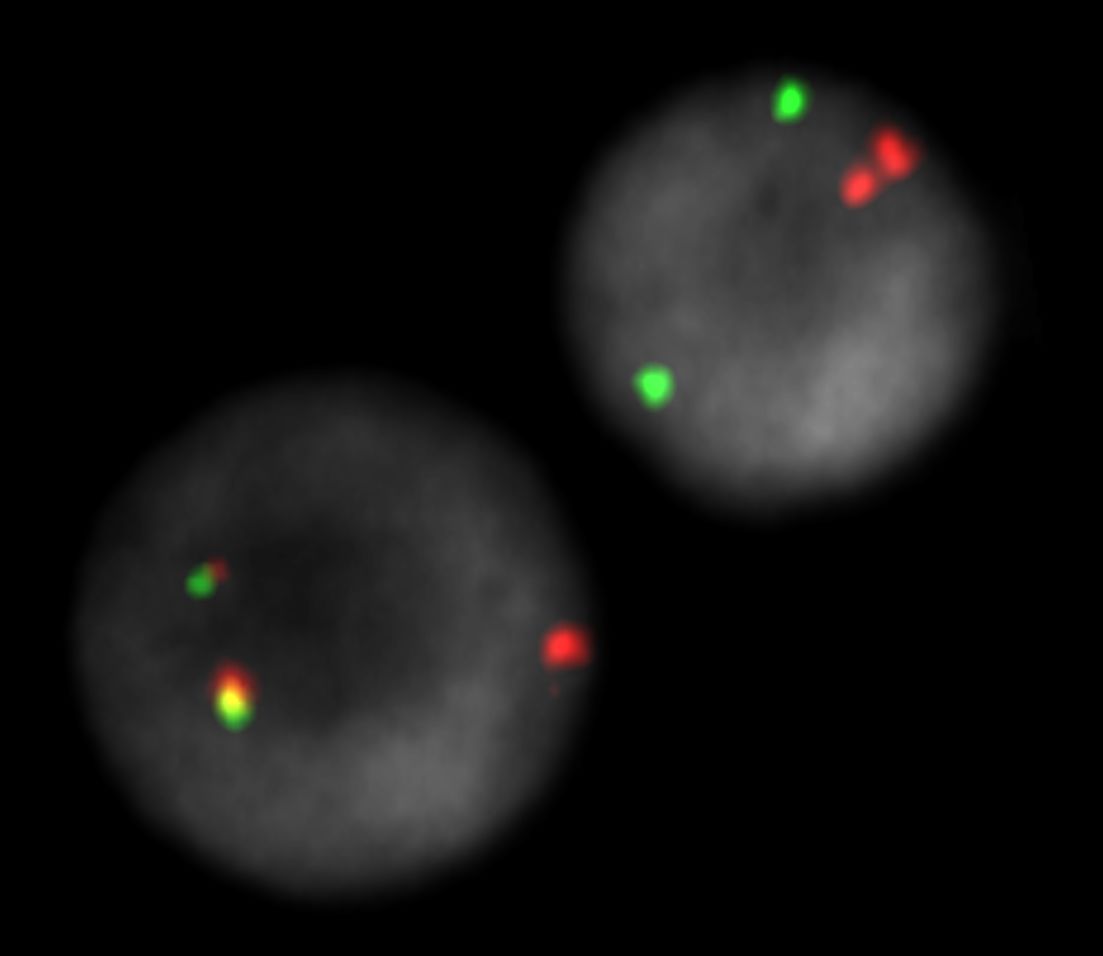
XL t(12;21) ETV6/RUNX1 DF hybridized to bone marrow cells. One normal interphase and one aberrant interphases are shown. The reciprocal translocation is splitting the orange and green signals, resulting in two fusion signals on the relevant chromosomes. One green loci is deleted. The relevant normal remaining loci is indicated by one orange signal.

Normal Cell:
Two green (2G) and two orange (2O) signals.

Aberrant Cell (typical results):
One green (1G), one orange (1O), and two green-orange colocalization/fusion signals (2GO) resulting from a reciprocal translocation between the relevant loci.

Aberrant Cell (typical results):
No green (no G), one orange (1O), and two green-orange colocalization/fusion signals (2GO) resulting from a reciprocal translocation between the relevant loci and a deletion of the locus covered by the green probe.
- Romana et al (1995) Blood 85:3662-3670
- Al-Obaidi et al (2002) Leukemia 16:669-674
- Sun et al (2017) Oncotarget 8:35445-35459
Certificate of Analysis (CoA)
or go to CoA Database



